Nelson H. Morgon1 e Aguinaldo R. de Souza2
1Instituto de Química e Centro de Computação em Engenharia e Ciências – Universidade Estadual de Campinas, Campinas, SP.
2Faculdade de Ciências da Universidade Estadual Paulista, Bauru, SP.
When we are in a feverish state due to some illness, such as the flu or a simple cold, we use some medicine derived from a drug found in nature or produced in the laboratory. When we ingest this medicine the objective is to recover our health, however, some very interesting phenomena occur in our organism at the molecular level that attract the interest of researchers. The attention is focused mainly on the study of the interactions between the drugs we ingest and the biomolecules in our body such as proteins and DNA.

HSA protein crystals.
Source: (https://www.imperial.ac.uk/people/s.curry)
Among a very large number of proteins found in our body, one of them is essential in the transport, or pharmacokinetics, of drugs and other molecules of biological interest: the Human Serum Albumin, or HSA. This protein, produced in the liver, is the most abundant in the blood, and performs many vital functions in our body, such as regulating the acidity (pH) of our blood. Another important function of this protein is the transport of the hormones of the thyroid gland, known as T3 and T4. HSA is a globular, heart-shaped protein that has the property of binding to various molecules either of origin within our own body (endogenous) or external to it (exogenous). The study of the binding of molecules to the HSA protein is accomplished by obtaining small crystals containing both the protein and the drug.
From these crystals and the X-ray diffraction technique we can obtain valuable information about the orientation of a drug within the protein. This information is essential for the development of new drugs for the treatment of diseases and the relief of symptoms arising from diseases or syndromes. However, for some drugs, after interaction with the HSA protein, it is not possible, and in some cases impossible, to obtain these crystals. Therefore, other resources are used, such as computer simulation of these chemical structures. Thus, with the appearance, in recent decades, of increasingly faster computers with greater information storage capacity, and also with the development of increasingly intelligent programs, it is now possible to study increasingly complex systems. Among these problems is the study of the interactions between drugs and the HSA protein. These studies are known in the scientific community as Molecular Modeling. The main characteristic of these studies is the predictive character of electronic and molecular properties of systems involving, for example, the study of isolated properties of drugs and HSA protein, as well as their interactions.
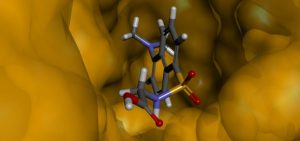
View of the cavity structure of the interaction site of HSA with the amino acid dansylglycine.
The mathematical calculations involved in these studies are relatively costly from a computational point of view, i.e., time consuming, and therefore our research group has been able to study the problems involving the interaction between different drugs and the HSA protein thanks to the infrastructure available at the Center for Computing in Engineering & Sciences (CCES). The results obtained in Molecular Modeling allow us to obtain information that can be used as subsidies in the interpretation of the results obtained experimentally (in laboratories), besides the possibility of predicting the chemical and physical properties of the drug-protein system. In the figure above, for example, we present one of the results obtained in the computational calculations, which shows a drug molecule inside the HSA protein.
Related scientific articles:
Venturini, D.; Pastrello, B.; Zeraik, M. L.; Pauli, I.; Andricopulo, A. D.; Silva-Filho, L. C.; Bolzani, V. S.; Morgon, N. H.; Souza, A. R.; Ximenes, V. F. Experimental, DFT and docking simulations of the binding of diapocynin to human serum albumin: induced circular dichroism. RSC Adv. 5(76), pp. 62220-62228, 2015.
De Souza, A. R.; Ximenes, V. F.; Morgon, N. H. Study of relative stability of tautomeric forms of phenylbutazone and calculation of UV-Vis and ECD spectra. Rev. Virtual. Quím. 8(2), pp. 525-535, 2016.
Venturini, D.; De Souza, A. R.; Caracelli, I.; Morgon, N. H.; SILVA-FILHO, L. C.; Ximenes, V. F. Induction of axial chirality in divanillin by interaction with bovine serum albumin. PLoS One, 12, pp. e0178597, 2017.
Ximenes, V. F.; Morgon, N. H., De Souza, A. R. Solvent-dependent inversion of circular dichroism signal in naproxen: An unusual effect!. Chirality 30, pp. 1049-1053, 2018.
De Souza, A. R.; Boza, I. A. F.; Ximenes, V. F.; Yoguim, M. I.; Dávila-Rodrigues, M. J. Morgon, N. H.; Caracelli, I. Elucidação da quiralidade induzida na molécula dansilglicina na complexação com a proteína albumina do soro humano (HSA). Quím. Nova, 42(2), pp. 135-142, 2019.
Sousa, I. L., Ximenes, V. F.; De Souza, A. R., Morgon, N. H. Solvent-Induced Stokes’ Shift in DCJTB: Experimental and Theoretical Results. J. Mol. Struct. (in press).

