This repository contains a set of source codes and input scripts that allows to perform nonequilibrium f
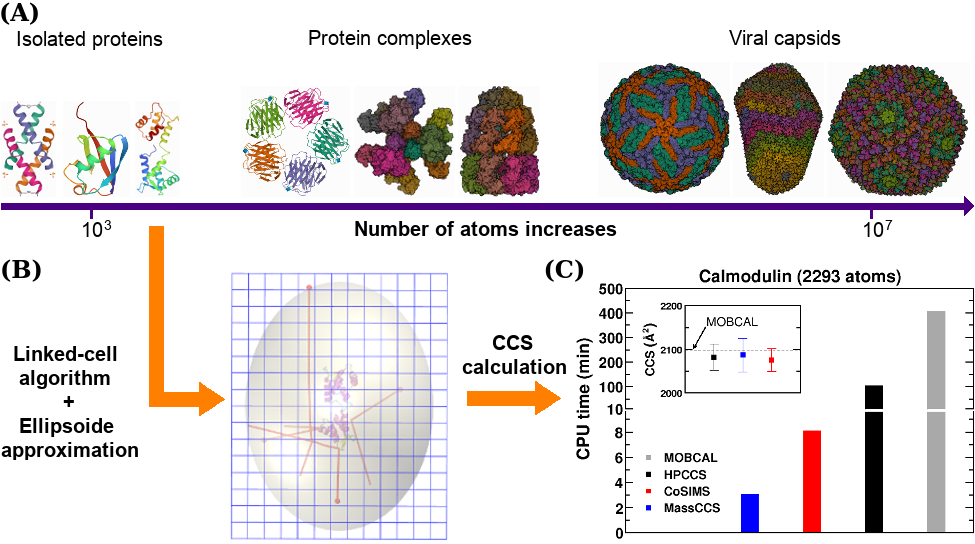
MassCCS is an efficient parallel software for calculating Collision Cross Section (CCS) under the trajectory method (TM) for any macromolecular structure regardless of its size, shape and surface rugosity, without significant loss of accuracy or performance.
MassCCS relies on the linked-cell algorithm for computing intermolecular forces, ellipsoid projection approximation and efficient parallelization techniques that greatly enhance its performance in comparison to available TM CCS codes.
MassCCS is written in C++ and supports OpenMP. Extensive tests on calculation accuracy, speed up gains, and scalability with system size were performed. MassCCS efficiency is particularly scalable for multiple CPU cores.
https://github.com/cces-cepid/massccs