Date: 19/02/2019, 14:00
Candidate: Guilherme Z M Fahur Bottino
Advisor: Prof. Leandro Martínez
Room IQ-03, Institute of Chemistry, University of Campinas
Abstract:
The problem of computational biomolecular modeling is one of the major themes of bioinformatics in the 21st century, and knowledge-based ab initio modeling, in particular, is the methodology of choice for the study of bleeding-edge problems, such as proteins that present little homology in the databases of known structures. An interesting way to counteract some drawbacks on this type of modeling is to assist it by 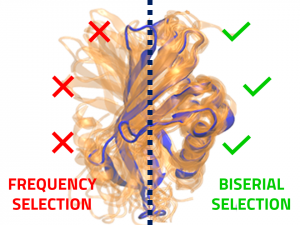 providing instrumental data on the protein structure in solution, and one of the experiments that can be drawn for this purpose is called Cross-Linking Mass Spectrometry, a technique that evaluates surface topological distance constraints. This technique can generate a large amount of data, of which only a small portion effectively represents the native constraints, giving rise to the challenge hence explored of selecting and recovering the appropriate constraints to provide as input to computational algorithms.
providing instrumental data on the protein structure in solution, and one of the experiments that can be drawn for this purpose is called Cross-Linking Mass Spectrometry, a technique that evaluates surface topological distance constraints. This technique can generate a large amount of data, of which only a small portion effectively represents the native constraints, giving rise to the challenge hence explored of selecting and recovering the appropriate constraints to provide as input to computational algorithms.
In the present work, we introduce a constraint score based on a classic psychometric quality indicator, called point-biserial correlation coefficient. We show, for different systems, that in almost all cases, properly applied biserial coefficient allows for the retrieval of more discriminating and informative constraints in relation to a given reference model. Successive constraint retrivals, over an iterative process, introduces incremental biases in the modeled sets to the point of shifting the sampled conformational space towards regions closer to what is believed to be the correct conformation, providing a general increase in modeling quality.
We show that, given an adequate number of recovered constraints and a good model selection tool, the constraints retrieved through BISCORE allow for significant increase in the amount of satisfactory models. Through a protocol that employs this methodology, implemented in a software package developed in this project, it was possible in some cases to increase the number of successful models by a 50-fold, when compared to preliminary unconstrained modeling.

